mne.compute_source_morph#
- mne.compute_source_morph(src, subject_from=None, subject_to='fsaverage', subjects_dir=None, zooms='auto', niter_affine=(100, 100, 10), niter_sdr=(5, 5, 3), spacing=5, smooth=None, warn=True, xhemi=False, sparse=False, src_to=None, precompute=False, verbose=None)[source]#
Create a SourceMorph from one subject to another.
Method is based on spherical morphing by FreeSurfer for surface cortical estimates [1] and Symmetric Diffeomorphic Registration for volumic data [2].
- Parameters:
- srcinstance of
SourceSpaces| instance ofSourceEstimate The SourceSpaces of subject_from (can be a SourceEstimate if only using a surface source space).
- subject_from
str|None Name of the original subject as named in the SUBJECTS_DIR. If None (default), then
src[0]['subject_his_id]'will be used.- subject_to
str|None Name of the subject to which to morph as named in the SUBJECTS_DIR. Default is
'fsaverage'. If None,src_to[0]['subject_his_id']will be used.Changed in version 0.20: Support for subject_to=None.
- subjects_dirpath-like |
None The path to the directory containing the FreeSurfer subjects reconstructions. If
None, defaults to theSUBJECTS_DIRenvironment variable.- zooms
float|tuple|str|None The voxel size of volume for each spatial dimension in mm. If spacing is None, MRIs won’t be resliced, and both volumes must have the same number of spatial dimensions. Can also be
'auto'to use5.ifsrc_to is Noneand the zooms fromsrc_tootherwise.Changed in version 0.20: Support for ‘auto’ mode.
- niter_affine
tupleofint Number of levels (
len(niter_affine)) and number of iterations per level - for each successive stage of iterative refinement - to perform the affine transform. Default is niter_affine=(100, 100, 10).- niter_sdr
tupleofint Number of levels (
len(niter_sdr)) and number of iterations per level - for each successive stage of iterative refinement - to perform the Symmetric Diffeomorphic Registration (sdr) transform. Default is niter_sdr=(5, 5, 3).- spacing
int|list|None The resolution of the icosahedral mesh (typically 5). If None, all vertices will be used (potentially filling the surface). If a list, then values will be morphed to the set of vertices specified in in
spacing[0]andspacing[1]. This will be ignored ifsrc_tois supplied.Changed in version 0.21: src_to, if provided, takes precedence.
- smooth
int|str|None Number of iterations for the smoothing of the surface data. If None, smooth is automatically defined to fill the surface with non-zero values. Can also be
'nearest'to use the nearest vertices on the surface.Changed in version 0.20: Added support for ‘nearest’.
- warnbool
If True, warn if not all vertices were used. The default is warn=True.
- xhemibool
Morph across hemisphere. Currently only implemented for
subject_to == subject_from. See notes below. The default is xhemi=False.- sparsebool
Morph as a sparse source estimate. Works only with (Vector) SourceEstimate. If True the only parameters used are subject_to and subject_from, and spacing has to be None. Default is sparse=False.
- src_toinstance of
SourceSpaces|None The destination source space.
For surface-based morphing, this is the preferred over
spacingfor providing the vertices.For volumetric morphing, this should be passed so that 1) the resultingmorph volume is properly constrained to the brain volume, and 2) STCs from multiple subjects morphed to the same destination subject/source space have the vertices.
For mixed (surface + volume) morphing, this is required.
New in v0.20.
- precomputebool
If True (default False), compute the sparse matrix representation of the volumetric morph (if present). This takes a long time to compute, but can make morphs faster when thousands of points are used. See
mne.SourceMorph.compute_vol_morph_mat()(which can be called later if desired) for more information.New in v0.22.
- verbosebool |
str|int|None Control verbosity of the logging output. If
None, use the default verbosity level. See the logging documentation andmne.verbose()for details. Should only be passed as a keyword argument.
- srcinstance of
- Returns:
- morphinstance of
SourceMorph The
mne.SourceMorphobject.
- morphinstance of
Notes
This function can be used to morph surface data between hemispheres by setting
xhemi=True. The full cross-hemisphere morph matrix maps left to right and right to left. A matrix for cross-mapping only one hemisphere can be constructed by specifying the appropriate vertices, for example, to map the right hemisphere to the left:vertices_from=[[], vert_rh], vertices_to=[vert_lh, []]
Cross-hemisphere mapping requires appropriate
sphere.left_rightmorph-maps in the subject’s directory. These morph maps are included with thefsaverage_symFreeSurfer subject, and can be created for other subjects with themris_left_right_registerFreeSurfer command. Thefsaverage_symsubject is included with FreeSurfer > 5.1 and can be obtained as described here. For statistical comparisons between hemispheres, use of the symmetricfsaverage_symmodel is recommended to minimize bias [1].New in v0.17.0.
New in v0.21.0: Support for morphing mixed source estimates.
References
Examples using mne.compute_source_morph#
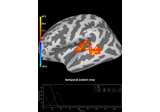
Permutation t-test on source data with spatio-temporal clustering
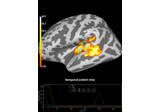
2 samples permutation test on source data with spatio-temporal clustering
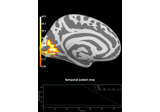
Repeated measures ANOVA on source data with spatio-temporal clustering

Compute sparse inverse solution with mixed norm: MxNE and irMxNE